Due: Thursday, April 18
Write a Perl program, webblast.pl, that uses the LPW module to access the BLAST server at the National Center for Biotechnology Information (NCBI) at http://www.ncbi.nlm.nih.gov/BLAST/. Your program should prompt the user for the name of an input file that contains a sequence nucleotides in FASTA format. (The book's file, record.fasta, is such a file. But you should probably review your notes to determine the syntax of FASTA files.) Your program will submit a request to the BLAST server via their web page and retrieve the resulting information.
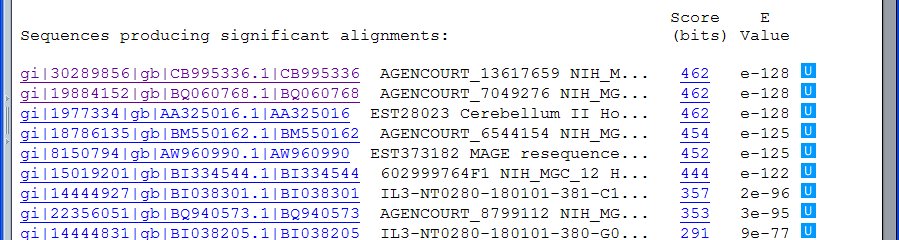
Here is the pertinent data from the BLAST server's response page:

Because the data (the FASTA sequence) you are transmitting to the server can be long, you must use a POST rather than a GET. If you carefully examine the html source code for the NCBI web page, you'll see that there is no explicit definitions of the names of the POST arguments. To determine what they are, make a local copy of the NCBI web page and alter it so that it uses a GET rather than a POST. When you launch the request from this page, carefully examine the URL and you should see the names of the arguments there.
The retrieval will be a bit tricky because the BLAST server usually needs about a minute to complete the query. Consequently, your program will have to be careful about how it submits its subsequent requests. You should print a message indicating approximately how long it will take for the request to be fulfilled. Your program should print a "still waiting" message to the console every 30 seconds to indicate the program has not frozen.
Your program should print the ten highest ranking alignments along with their scores. Here is a sample output:
Score E
Sequences producing significant alignments: (bits) Value Identities Gaps
AGENCOURT_13617659 NIH_MGC_148 Homo sapiens cDNA clone 462 e-128 240/241 (99%) 1/241 (0%)
AGENCOURT_7049276 NIH_MGC_99 Homo sapiens cDNA clone 462 e-128 240/241 (99%) 1/241 (0%)
...
Note that the names of the sequences, and the Identies and Gaps information can be obtained by following links from the retrieved document (in the diagram above, the links start with "gi|") or by a more complex parsing of the retrieved document. Note, too, that you may have to calculate the value for the Gaps field, as it is not always present in the server's output.